Hello and welcome! We are a biomolecular NMR lab studying the importance of protein dynamics in function and malfunction. Please feel free to explore this website to learn more about our research, as well as the people who are carrying it out.
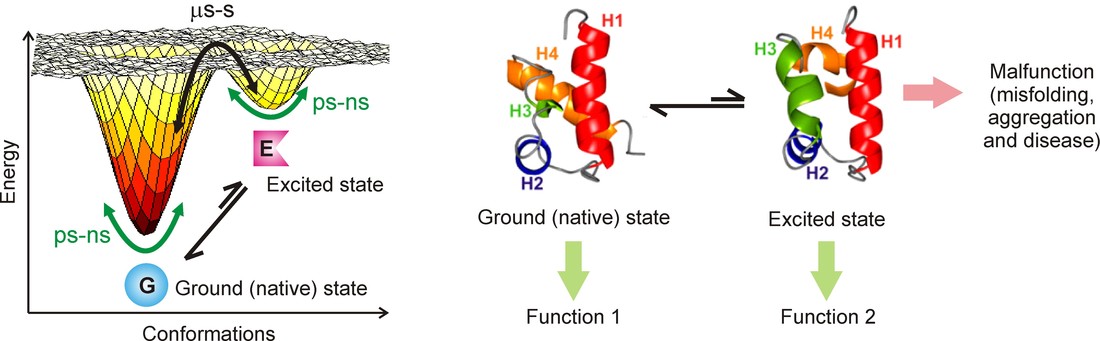
The canonical sequence-structure-function paradigm in molecular biology states that the amino acid sequence of a protein codes for a unique three-dimensional structure that is necessary for the protein to perform its function. However, recent research has shown that the picture of a protein as a single static native state structure is incomplete. Proteins exist not as isolated molecules, but reside instead in a water bath at a certain temperature; the thermal energy of these solvent molecules is transferred to the protein via collisions, resulting in changes or fluctuations in the protein structure. Because of thermal energy, proteins can sample higher energy states (conformationally excited, or just excited states) with structures very different from the ground native state. It follows in turn that these excited states can adopt functions distinct from the ground state, and that the complete functional space of a protein can be elucidated only through a knowledge of all the thermally accessible structures of the protein and not merely the native state structure. In addition, these excited states can be more aggregation-prone than the native state, causing downstream cellular malfunction and disease.
NMR spectroscopy is a very powerful technique for probing the structure and dynamics of proteins (Why NMR?). We use the information from NMR spectroscopy, supplemented with data from other biophysical techniques, to understand how the fluctuations in the structure of proteins participating in a variety of cellular processes help in their function, or result in their malfunction. For more details on current projects, please visit our research page here.